Low-pass sequencing and imputation for evaluating genetic
By A Mystery Man Writer
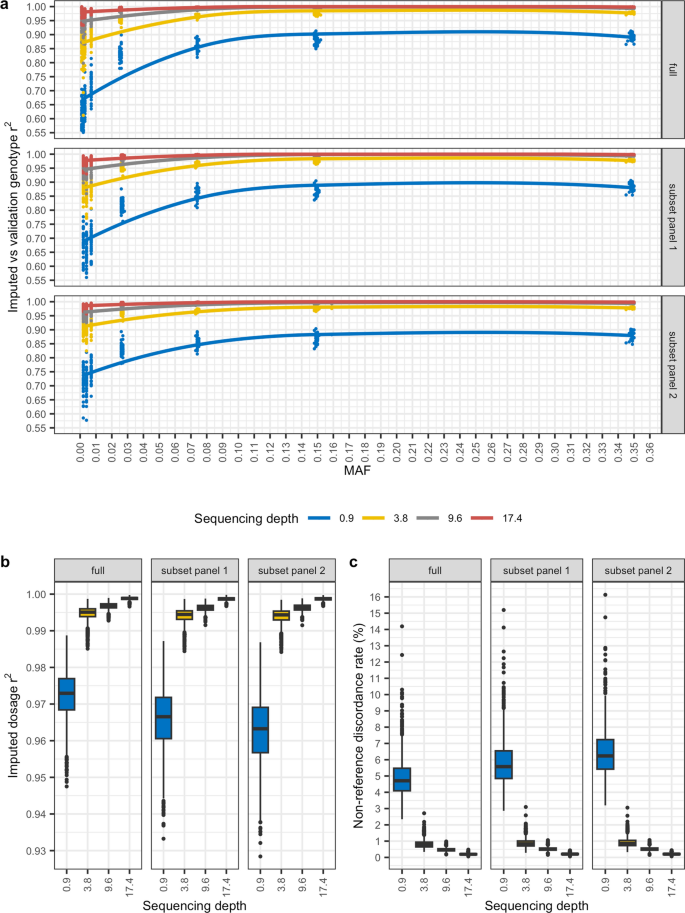
A cautionary tale of low-pass sequencing and imputation with respect to haplotype accuracy, Genetics Selection Evolution
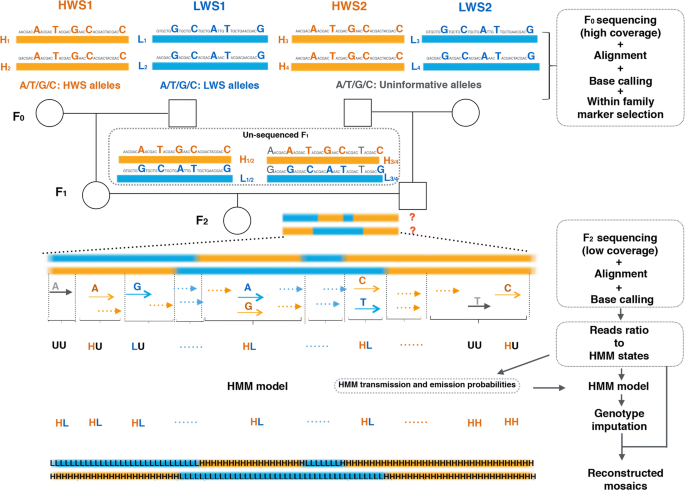
Genotyping by low-coverage whole-genome sequencing in intercross pedigrees from outbred founders: a cost-efficient approach, Genetics Selection Evolution
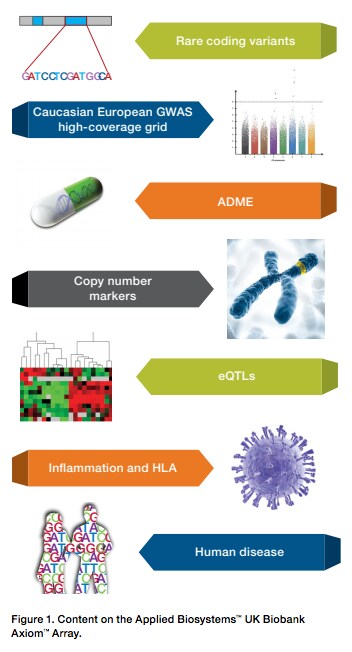
Biobanks and Beyond: Genotyping for the Future

MagicalRsq: Machine-learning-based genotype imputation quality calibration - ScienceDirect

International Society for Forensic Genetics 2022

Evaluating genotype imputation pipeline for ultra-low coverage ancient genomes
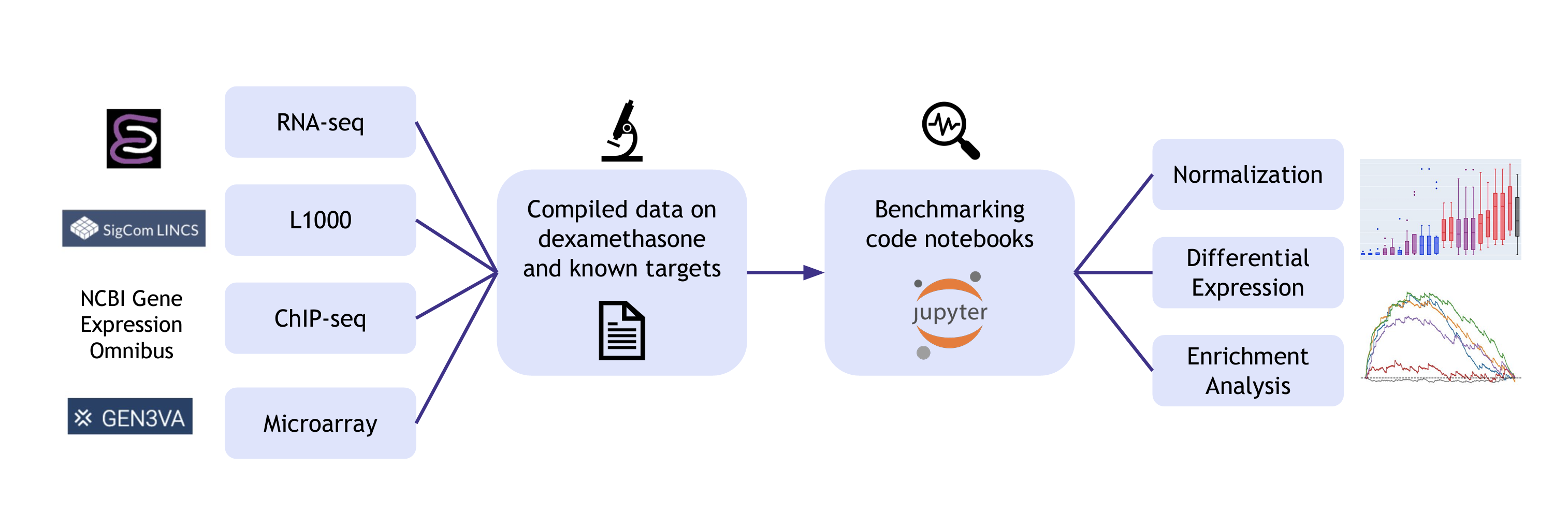
The Dex-Benchmark resource – datasets and code to evaluate algorithms for transcriptomics data analysis
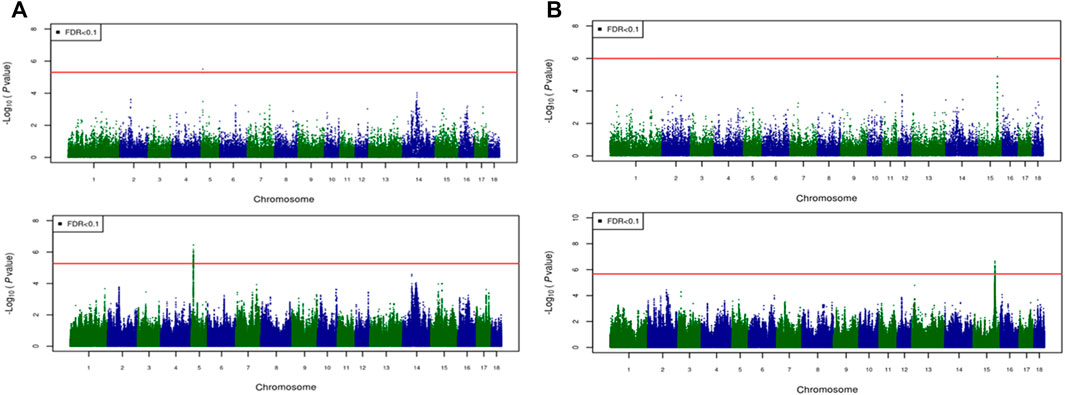
Frontiers Imputation to whole-genome sequence and its use in genome-wide association studies for pork colour traits in crossbred and purebred pigs
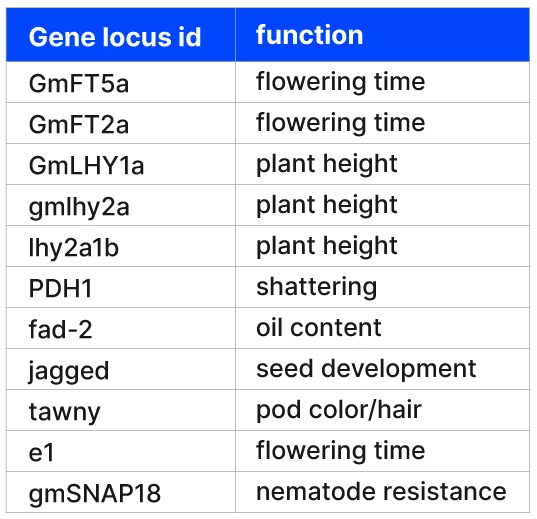
Exploring the power of low-pass WGS with target capture in plant breeding - Gencove
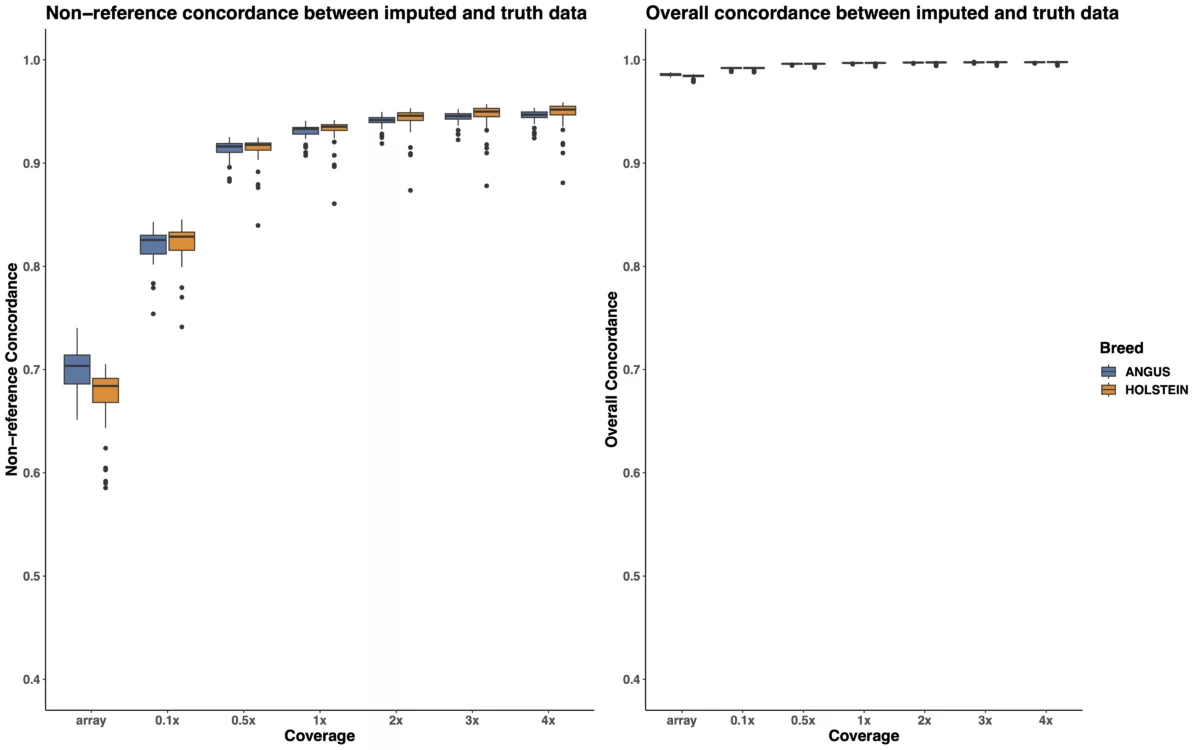
Low pass sequencing plus imputation in cattle outperforms imputation from genotyping arrays - Gencove
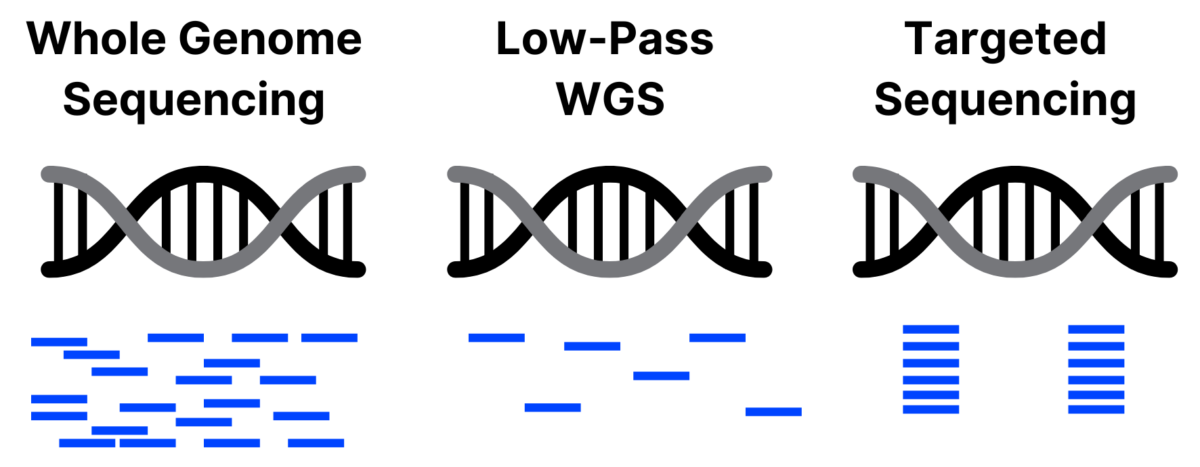
Low-Pass Whole Genome Sequencing

Genotype imputation in a sample of apparently unrelated individuals

Frontiers Ultra Low-Coverage Whole-Genome Sequencing as an Alternative to Genotyping Arrays in Genome-Wide Association Studies
- PDF) A beginner's guide to low-coverage whole genome sequencing for population genomics
- Site Selection for Land-Intensive Occupiers in Infill Areas — Chuck Berger

- Women's Low Coverage Mini Bralette Bikini Top - Wild Fable™ Black XXS
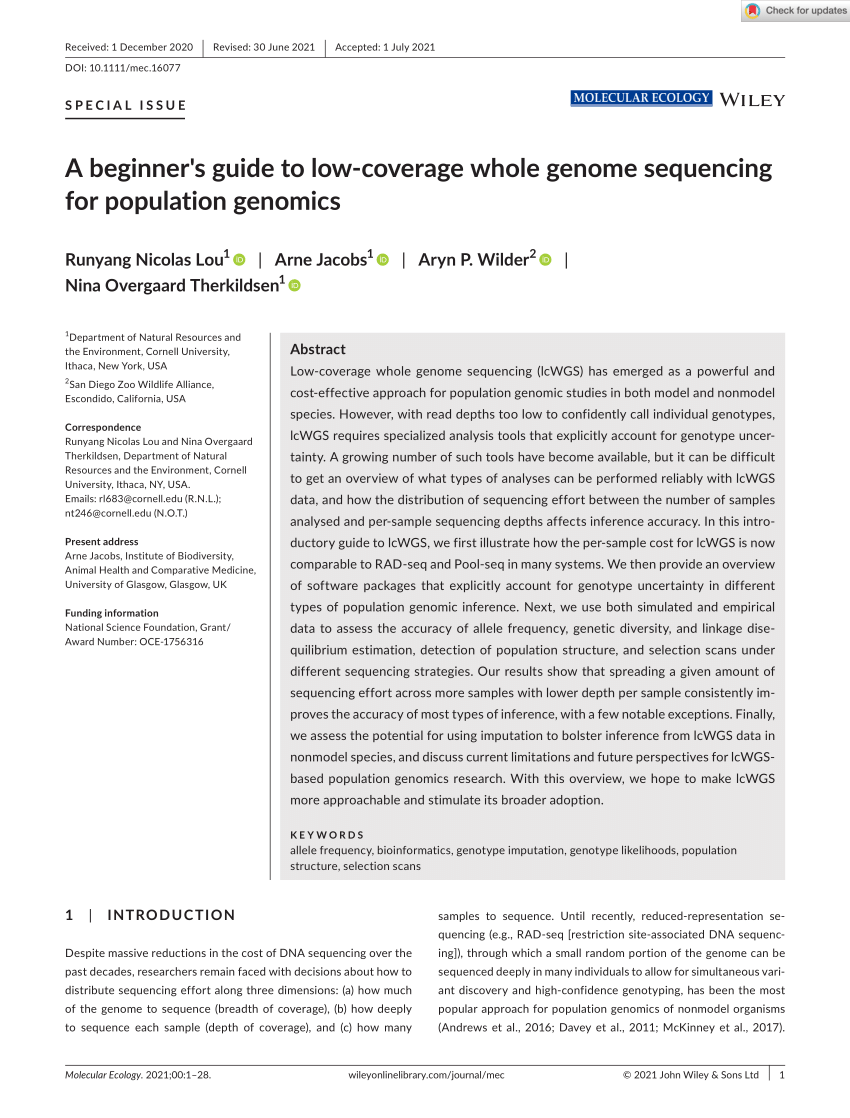
- School-Based Interventions to Increase Student COVID-19 Vaccination Coverage in Public School Populations with Low Coverage — Seattle, Washington, December 2021–June 2022
- How to Purchase Contractors General Liability Insurance
- American Girl Wellie Wisher Camille Doll, doll clothing and books
- Lingerie Romance Favorita G e M - Roupas - Betânia, Manaus

- Elevate Leggings – IRONGEAR Fitness

- Envie Women's Cotton Minimizer Bra/Non-Padded, Wirefree, Cut Seam Bra/Inner Wear for Ladies Daily Use Bra

- Mother Of The Bride Pant Suits Plus Size Elegant Women Long


