A protocol for applying low-coverage whole-genome sequencing data
By A Mystery Man Writer
STAR Protocols is an open access, peer-reviewed journal from Cell Press. We offer structured, transparent, accessible, and repeatable step-by-step experimental and computational protocols from all areas of life, health, earth and physical sciences.

Using low-coverage whole genome sequencing (genome skimming) to delineate three introgressed species of buffalofish (Ictiobus) - ScienceDirect
Shuhua-Group · GitHub

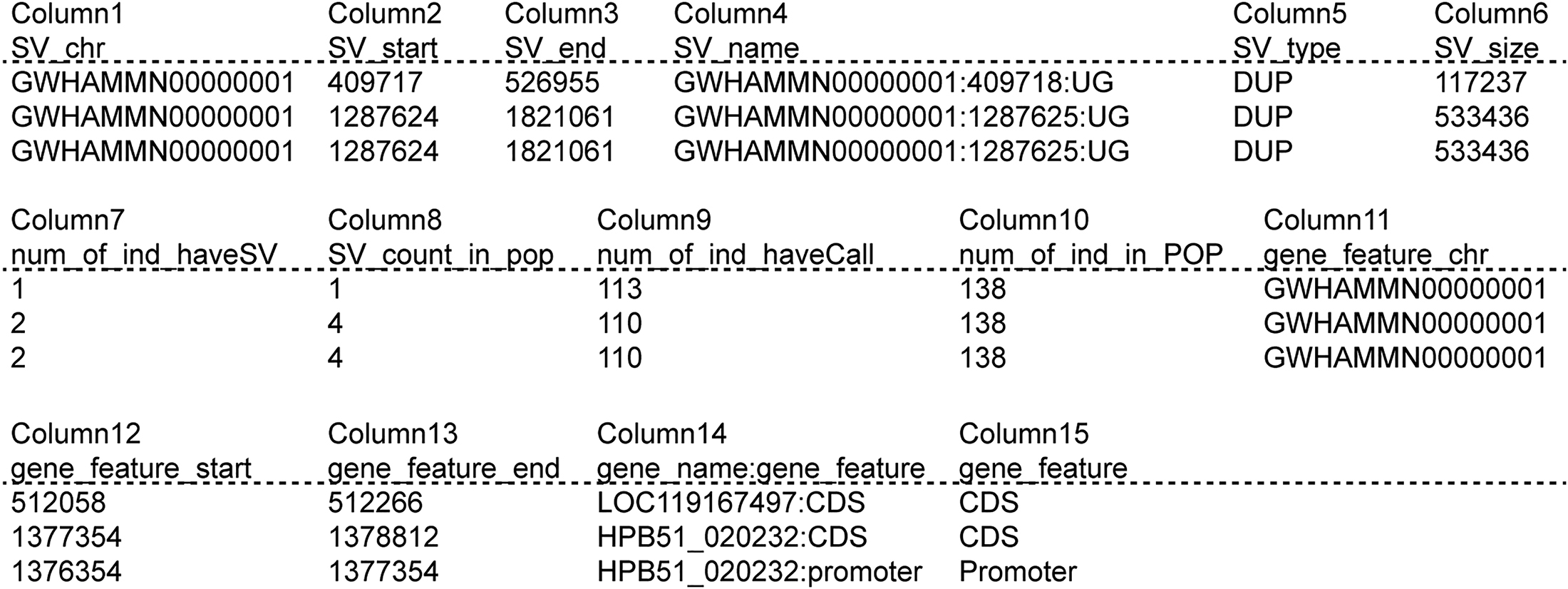
PDF) Mapping structural variations in and reveals vector–pathogen adaptation
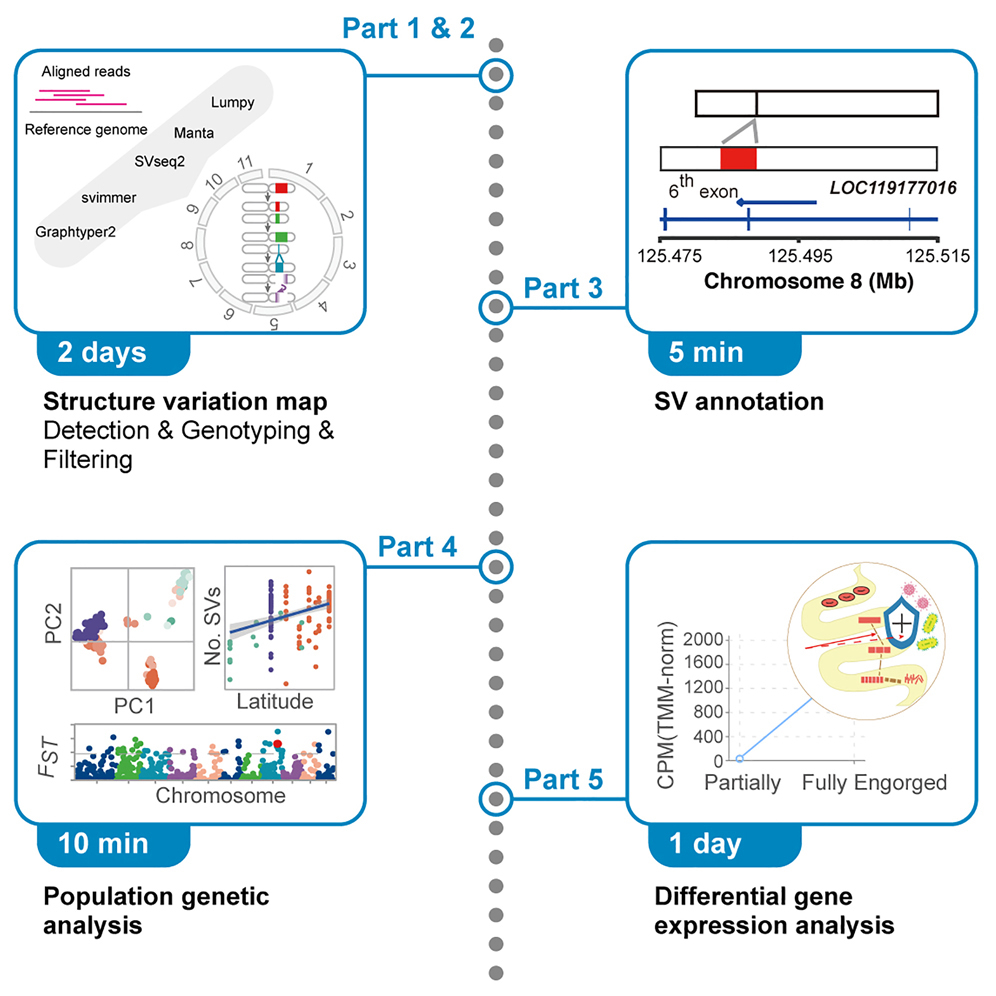
A protocol for applying low-coverage whole-genome sequencing data in structural variation studies

PDF) A protocol for applying low-coverage whole-genome sequencing data in structural variation studies
GitHub - Shuhua-Group/SVanalysis_STARProtocols: A protocol for applying low-coverage whole-genome sequencing data in structure variation studies

PDF) PGG.SV: a whole-genome-sequencing-based structural variant resource and data analysis platform

A complete protocol for whole-genome sequencing of virus from clinical samples: Application to coronavirus OC43 - ScienceDirect
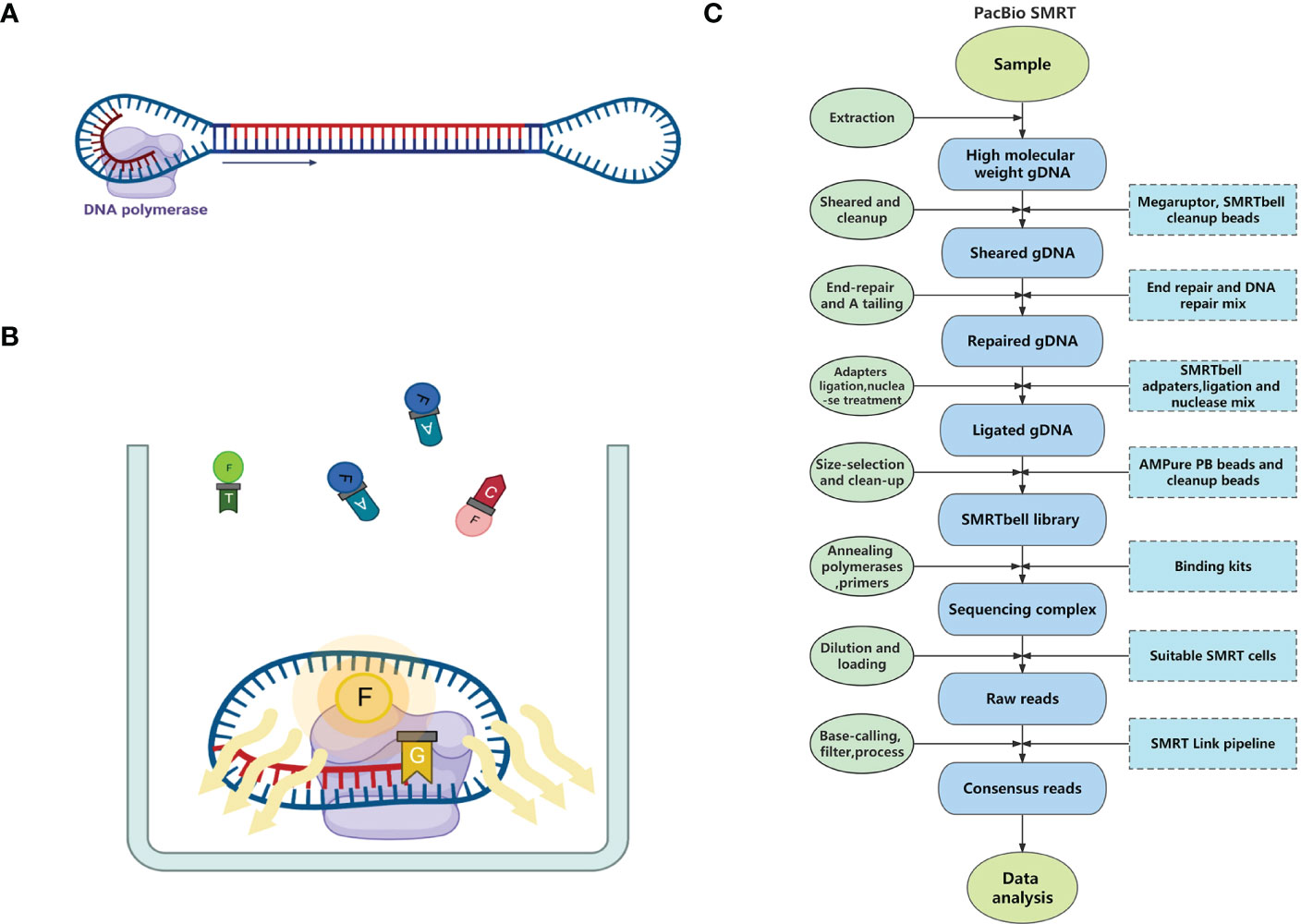
Frontiers Application of third-generation sequencing to herbal genomics

PDF] A spectrum of free software tools for processing the VCF variant call format: vcflib, bio-vcf, cyvcf2, hts-nim and slivar
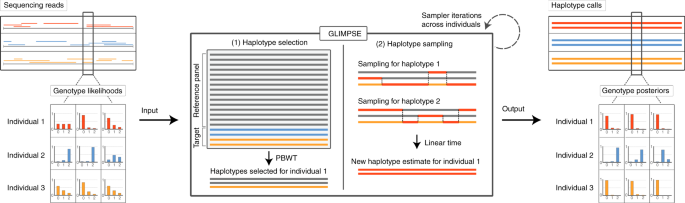
Efficient phasing and imputation of low-coverage sequencing data using large reference panels
- Network Coverage Icon Low Network Coverage Stock Illustration 2104420790

- Capsule network-based approach for estimating grassland coverage using time series data from enhanced vegetation index - ScienceDirect

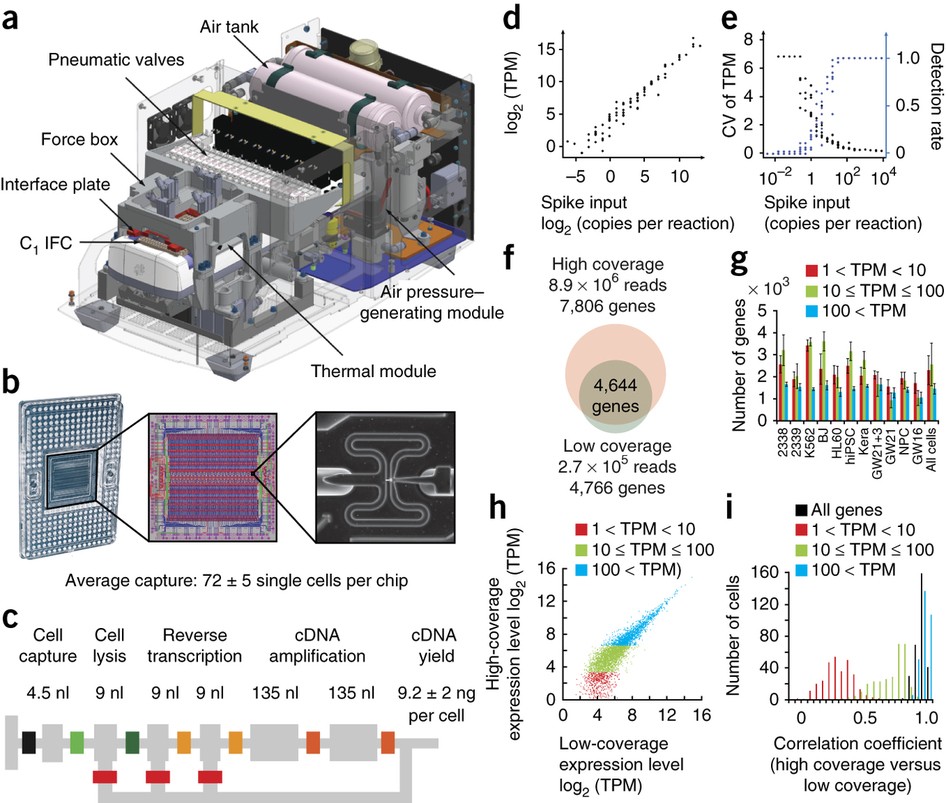
- Low-coverage single-cell mRNA sequencing reveals cellular heterogeneity and activated signaling pathways in developing cerebral cortex
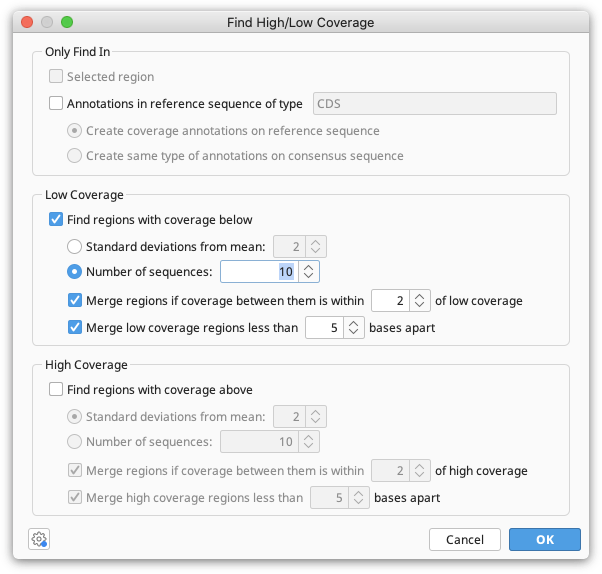
- Assembly of SARS-CoV-2 genomes from tiled amplicon Illumina sequencing using Geneious Prime – Geneious
- Women's Low Coverage Mini Triangle Bikini Top - Wild Fable™ Black XXS





