Efficient phasing and imputation of low-coverage sequencing data
By A Mystery Man Writer

PDF) Publisher Correction: Efficient phasing and imputation of low

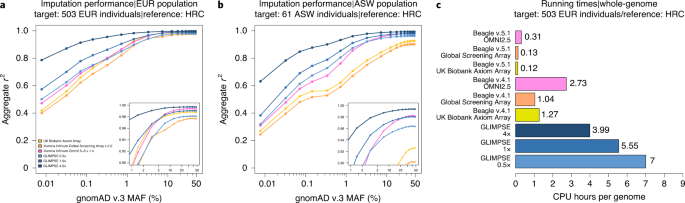
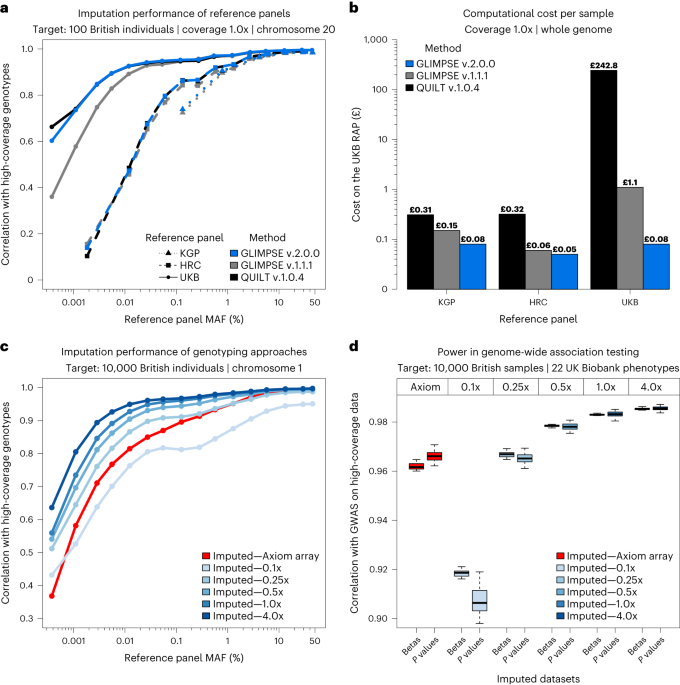
Imputation of low-coverage sequencing data from 150,119 UK Biobank
Frontiers Comparison of Genotype Imputation for SNP Array and Low-Coverage Whole-Genome Sequencing Data

Frontiers Ultra Low-Coverage Whole-Genome Sequencing as an

Low-coverage sequencing cost-effectively detects known and novel variation in underrepresented populations - ScienceDirect

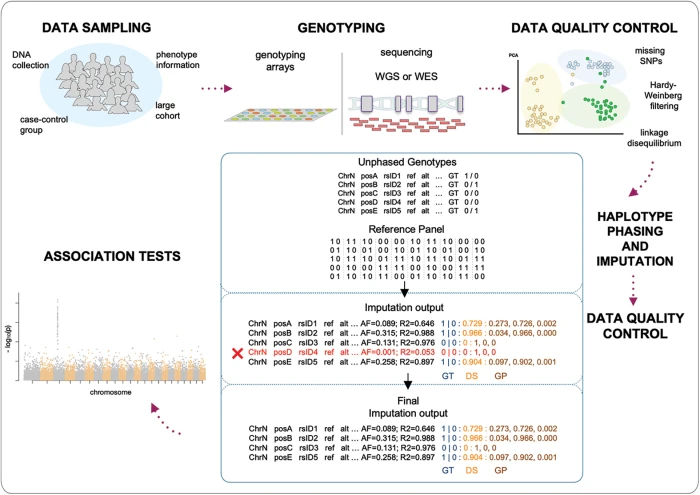
Assessment of the performance of different imputation methods for

Phasing performance. (a,b) Switch error rate (SER, y-axis, log

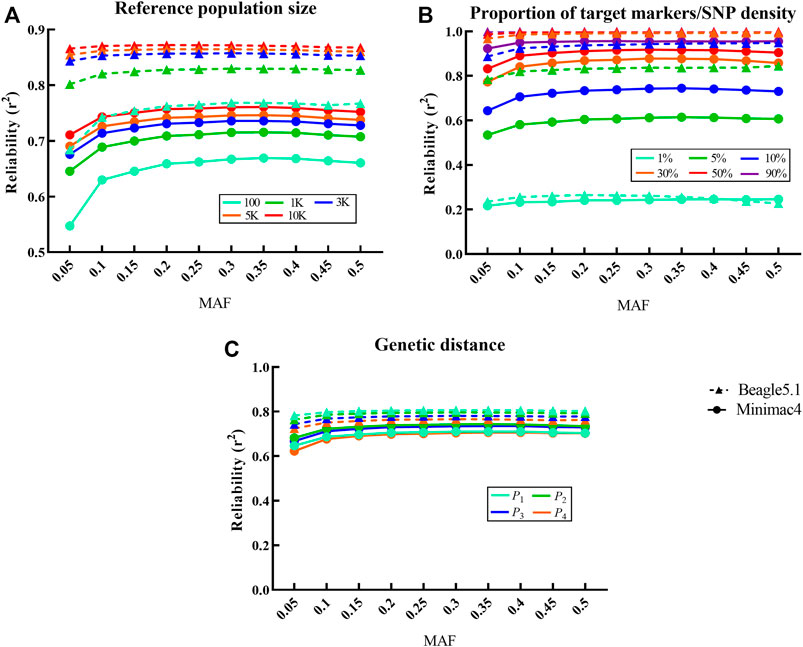
A comparative analysis of current phasing and imputation software

The data behind increasing adoption of low-pass sequencing in human genomics, by Joe Pickrell, The Gencove Blog

Evaluation of low-pass genome sequencing in polygenic risk score

Accurate rare variant phasing of whole-genome and whole-exome sequencing data in the UK Biobank - Abstract - Europe PMC

Animals, Free Full-Text
BIOCELL, Free Full-Text

Schematic of the QUILT model The model for one Gibbs sampling is
- Imputation of low-coverage sequencing data from 150,119 UK Biobank genomes
- How to improve the Wi-Fi® or wireless internet connection to your Roku® streaming device

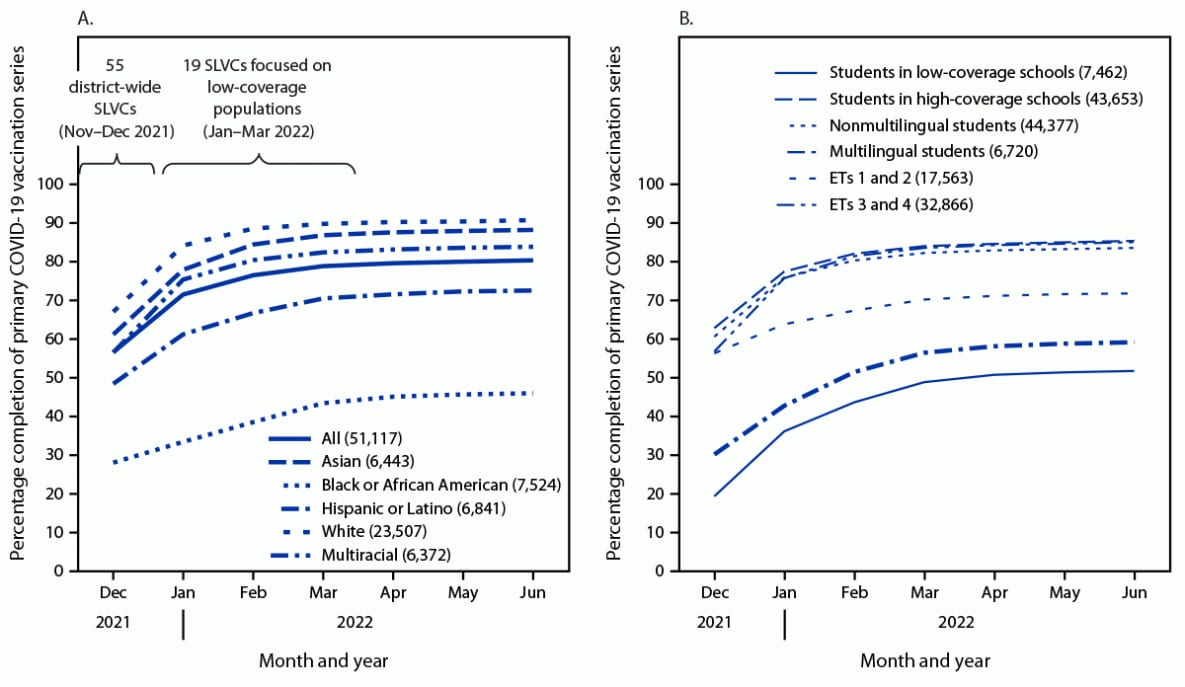
- School-Based Interventions to Increase Student COVID-19 Vaccination Coverage in Public School Populations with Low Coverage — Seattle, Washington, December 2021–June 2022
- Network Coverage Icon Low Network Coverage Stock Illustration

- Jacoco reports low coverage with Compose tests · Issue #1208
- NWT Lululemon Define Jacket *Nulu Size 10 Strawberry Milkshake -LW4CFOS STMI

- Burning Man wedding ceremony and the story about how I've lost my

- LULULEMON SHORTS HAUL REVIEW / SPEED UP LOW-RISE SHORT + TRACKER

- Feel Like Skin Yoga Leggings in 7 Color Choices in Size 4, 6, 8

- Anti Glare Glasses Night Vision Goggles for Driving Sight Goggles - Nightzer™
